Fully Convolutional Networks for Cardiac Image Segmentation
This is a neural network method which segments a cardiac MR image sequence across all the time frames of a cardiac cycle. The network was trained on the UK Biobank cardiac image set and it achieves a segmentation performance comparable to human experts. The following movies illustrate the segmentation results on short-axis and long-axis cine images. The source code and the trained model can be downloaded at [github].
(a) Short-axis image (basal).
(b) Short-axis image (mid-ventricular).
(c) Short-axis image (apical).
(d) Long-axis image (2-chamber view).
(e) Long-axis image (4-chamber view).
Related publications:
1. W. Bai, M. Sinclair, G. Tarroni, O. Oktay, M. Rajchl, G. Vaillant, A.M. Lee, N. Aung, E. Lukaschuk, M.M. Sanghvi, F. Zemrak, K. Fung, J.M. Paiva, V. Carapella, Y.J. Kim, H. Suzuki, B. Kainz, P.M. Matthews, S.E. Petersen, S.K. Piechnik, S. Neubauer, B. Glocker and D. Rueckert. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Journal of Cardiovascular Magnetic Resonance, accepted.
Cardiac Image Multi-Atlas Segmentation Pipeline
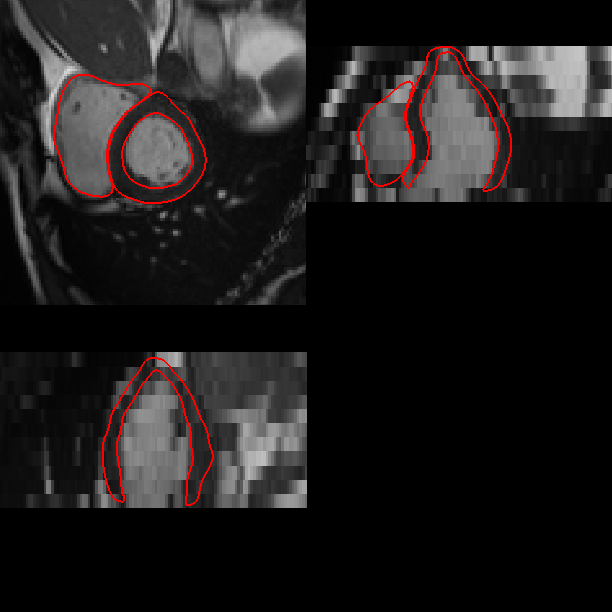
This is a software pipeline which segments a cardiac MR image sequence at two time frames, end-diastole (ED) and end-systole (ES) using multi-atlas segmentation method. Given an input image (a), the pipeline calls image registration and label fusion programmes and produces segmentation and meshes as shown in (b) and (c). The pipeline can be downloaded at [github].
(a) A cardiac MR image at end-diastole.

(b) Segmentation result.
(c) Fitted meshes.
Related publications:
1. W. Bai, W. Shi, C. Ledig and D. Rueckert. Multi-atlas segmentation with augmented features for cardiac MR images. Medical Image Analysis, 19(1):98-109, 2015.
2. W. Bai, W. Shi, A. de Marvao, T.J.W. Dawes, D.P. O’Regan, S.A. Cook, D. Rueckert. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Medical Image Analysis, 26(1):133-145, 2015.
3. W. Bai, W. Shi, D.P. O’Regan, T. Tong, H. Wang, S. Jamil-Copley, N.S. Peters and D. Rueckert. A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: Application to cardiac MR images. IEEE Transactions on Medical Imaging, 32(7):1302-1315, 2013.
4. O. Oktay, W. Bai, R. Guerrero, M. Rajchl, A. de Marvao, D. O’Regan, S. Cook, M. Heinrich, B. Glocker, and D. Rueckert. Stratified decision forests for accurate anatomical landmark localization in cardiac images. IEEE Transactions on Medical Imaging, 36(1):332-342, 2017.
Multi-Atlas Label Fusion
The label_fusion programme is just one step in the multi-atlas segmentation pipeline. It is already installed if you have installed the above segmentation pipeline. It can also be used separately with other image registration tools. The programme can be downloaded at this [link].
Currently, the label fusion programme includes four algorithms:
1. majority-vote (MV)
2. patch-based segmentation (PB)
3. patch-based segmentation with augmented features (PBAF)
4. SVM segmentation with augmented features (SVMAF)
A statically linked executable has already been provided. It was compiled in Ubuntu 12.04. To test the algorithms, please go to the demo_script directory and run the demo script on the testing data. You can also compile the code on your own. To that end, Prof. Daniel Rueckert’s [IRTK] needs to be downloaded and compiled at the first place.