Presentations
Since April 2020, the COVID pandemic has led to many international conferences being moved to virtual events. Our researchers at SmartHeart have given numerous virtual presentations over this time, and here we share a selection of their posters and talks.
E-posters
Presentations
Esther Anton-Puyol et al.
5 mins 36 secs
Abstract:
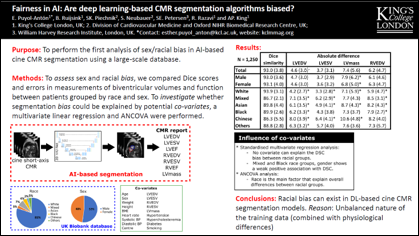
The subject of ‘fairness’ in artificial intelligence (AI) refers to assessing AI algorithms for potential bias based on demographic characteristics such as race and gender, and the development of algorithms to address this bias. Most applications to date have been in computer vision, although some work in healthcare has started to emerge. The use of deep learning (DL) in cardiac MR segmentation has led to impressive results in recent years, and such techniques are starting to be translated into clinical practice. However, no work has yet investigated the fairness of such models. In this work, we perform such an analysis for racial/gender groups, focusing on the problem of training data imbalance, using a nnU-Net model trained and evaluated on cine short axis cardiac MR data from the UK Biobank dataset, consisting of 5,903 subjects from 6 different racial groups. We find statistically significant differences in Dice performance between different racial groups. To reduce the racial bias, we investigated three strategies: (1) stratified batch sampling, in which batch sampling is stratified to ensure balance between racial groups; (2) fair meta-learning for segmentation, in which a DL classifier is trained to classify race and jointly optimized with the segmentation model; and (3) protected group models, in which a different segmentation model is trained for each racial group. We also compared the results to the scenario where we have a perfectly balanced database. To assess fairness we used the standard deviation (SD) and skewed error ratio (SER) of the average Dice values. Our results demonstrate that the racial bias results from the use of imbalanced training data, and that all proposed bias mitigation strategies improved fairness, with the best SD and SER resulting from the use of protected group models.
Generalised low-rank non-rigid motion corrected reconstruction for 3D free-breathing liver MRF
Gastao Cruz, Olivier Jaubert, Haikun Qi, Torben Schneider, Rene Botnar and Claudia Prieto
5 mins 8 secs
Abstract: Magnetic Resonance Fingerprinting (MRF) has been shown to enable simultaneous T1 and T2 mapping of the liver and abdomen. 2D liver MRF requires breath holding, whereas preliminary results have been demonstrated for 3D free-breathing liver MRF using respiratory gating. However, gating approaches lead to unpredictable scan times and may impair the MRF encoding, since only data within the respiratory gating window is used for reconstruction. Here we propose a novel low-rank motion corrected approach to both resolve MRF varying contrast and perform non-rigid respiratory motion correction directly in the reconstruction, enabling 3D free-breathing liver MRF with 100% respiratory scan efficiency.
Generalized low-rank non-rigid motion corrected reconstruction for 2D cardiac MRF
Gastao Cruz, Haikun Qi, Olivier Jaubert, Aurelien Bustin, Thomas Kuestner, Torben Schneider, Rene Botnar and Claudia Prieto
7 mins 49 secs
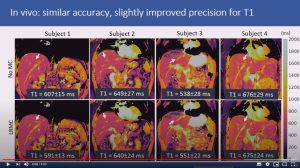
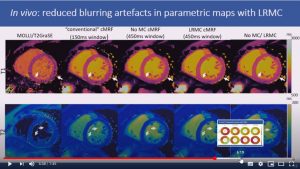
Abstract: Cardiac Magnetic Resonance Fingerprinting (cMRF) has been proposed for simultaneous myocardial T1 and T2 mapping. This approach uses ECG-triggering to synchronize data acquisition to a small mid-diastolic window, reducing cardiac motion artefacts but also limiting the amount of acquired data per heartbeat. This low scan efficiency can limit the spatial resolution achievable in a breath-held scan. Here we introduce a novel approach for contrast-resolved motion-corrected reconstruction, that combines the generalized matrix description formulism for non-rigid motion correction with low-rank compression of temporally varying contrast. This approach enables longer acquisition windows and higher scan efficiency in cMRF, correcting for cardiac motion.
Realistic Adversarial Data Augmentation for MR Image Segmentation
Chen Chen, Chen Qin, Huaqi Qiu, Cheng Ouyang, Shuo Wang, Liang Chen, Giacomo Tarroni, Wejia Bai and Daniel Rueckert
9 mins 43 secs
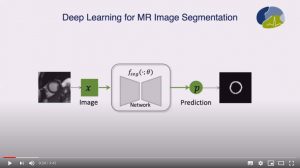
Abstract: Neural network-based approaches can achieve high accuracy in various medical image segmentation tasks. However, they generally require large labelled datasets for supervised learning. Acquiring and manually labelling a large medical dataset is expensive and sometimes impractical due to data sharing and privacy issues. In this work, we propose an adversarial data augmentation method for training neural networks for medical image segmentation. Instead of generating pixel-wise adversarial attacks, our model generates plausible and realistic signal corruptions, which models the intensity inhomogeneities caused by a common type of artefacts in MR imaging: bias field. The proposed method does not rely on generative networks, and can be used as a plug-in module for general segmentation networks in both supervised and semi-supervised learning. Using cardiac MR imaging we show that such an approach can improve the generalization ability and robustness of models as well as provide significant improvements in low-data scenarios.
Quality-aware semi-supervised learning for CMR segmentation
Bram Ruijsink
9 mins 53 secs
Abstract: One of the challenges in developing deep learning algorithms for medical image segmentation is the scarcity of annotated training data. To overcome this limitation, data augmentation and semi-supervised learning (SSL) methods have been developed. However, these methods have limited effectiveness as they either exploit the existing data set only (data augmentation) or risk negative impact by adding poor training examples (SSL). Segmentations are rarely the final product of medical image analysis – they are typically used in downstream tasks to infer higher-order patterns to evaluate diseases. Clinicians take into account a wealth of prior knowledge on biophysics and physiology when evaluating image analysis results. We have used these clinical assessments in previous works to create robust quality-control (QC) classifiers for automated cardiac magnetic resonance (CMR) analysis. In this paper, we propose a novel scheme that uses QC of the downstream task to identify high quality outputs of CMR segmentation networks, that are subsequently utilised for further network training. In essence, this provides quality-aware augmentation of training data in a variant of SSL for segmentation networks (semiQCSeg). We evaluate our approach in two CMR segmentation tasks (aortic and short axis cardiac volume segmentation) using UK Biobank data and two commonly used network architectures (U-net and a Fully Convolutional Network) and compare against supervised and SSL strategies. We show that semiQCSeg improves training of the segmentation networks. It decreases the need for labelled data, while outperforming the other methods in terms of Dice and clinical metrics. SemiQCSeg can be an efficient approach for training segmentation networks for medical image data when labelled datasets are scarce.
Interpretable Deep Models for Cardiac Resynchronisation Therapy Response Prediction
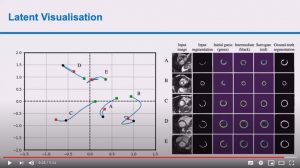
Esther Puyol-Antón, Chen Chen, James R. Clough, Bram Ruijsink, Baldeep S. Sidhu, Justin Gould, Bradley Porter, Marc Elliott, Vishal Mehta, Daniel Rueckert, Christopher A. Rinaldi, and Andrew P. King
9 mins 50 secs
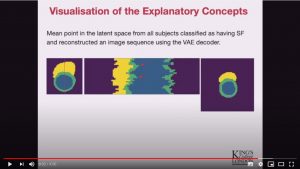
Abstract: Advances in deep learning (DL) have resulted in impressive accuracy in some medical image classification tasks, but often deep models lack interpretability. The ability of these models to explain their decisions is important for fostering clinical trust and facilitating clinical translation. Furthermore, for many problems in medicine there is a wealth of existing clinical knowledge to draw upon, which may be useful in generating explanations, but it is not obvious how this knowledge can be encoded into DL models – most models are learnt either from scratch or using transfer learning from a different domain. In this paper we address both of these issues. We propose a novel DL framework for image-based classification based on a variational autoencoder (VAE). The framework allows prediction of the output of interest from the latent space of the autoencoder, as well as visualisation (in the image domain) of the effects of crossing the decision boundary, thus enhancing the interpretability of the classifier. Our key contribution is that the VAE disentangles the latent space based on ‘explanations’ drawn from existing clinical knowledge. The framework can predict outputs as well as explanations for these outputs, and also raises the possibility of discovering new biomarkers that are separate (or disentangled) from the existing knowledge. We demonstrate our framework on the problem of predicting response of patients with cardiomyopathy to cardiac resynchronization therapy (CRT) from cine cardiac magnetic resonance images. The sensitivity and specificity of the proposed model on the task of CRT response prediction are 88.43% and 84.39% respectively, and we showcase the potential of our model in enhancing understanding of the factors contributing to CRT response.
Deep Generative Model-based Quality Control for Cardiac MRI Segmentation
Shuo Wang, Giacomo Tarroni, Chen Qin, Yuanhan Mo, Chengliang Dai, Chen Chen, Ben Glocker, YiKe Guo, Daniel Rueckert and Wenjia Bai
9 mins 54 secs
Abstract: In recent years, convolutional neural networks have demonstrated promising performance in a variety of medical image segmentation tasks. However, when a trained segmentation model is deployed into the real clinical world, the model may not perform optimally. A major challenge is the potential poor-quality segmentations generated due to degraded image quality or domain shift issues. There is a timely need to develop an automated quality control method that can detect poor segmentations and feedback to clinicians. Here we propose a novel deep generative model-based framework for quality control of cardiac MRI segmentation. It first learns a manifold of good-quality image-segmentation pairs using a generative model. The quality of a given test segmentation is then assessed by evaluating the difference from its projection onto the good-quality manifold. In particular, the projection is refined through iterative search in the latent space. The proposed method achieves high prediction accuracy on two publicly available cardiac MRI datasets. Moreover, it shows better generalisation ability than traditional regression-based methods. Our approach provides a real-time and model-agnostic quality control for cardiac MRI segmentation, which has the potential to be integrated into clinical image analysis workflows.
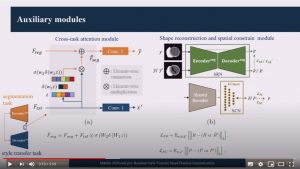
Random Style Transfer based domain Generalization Integrating Shape and Spatial Information
Lei Li, Veronika Zimmer, Wangbin Ding, Fuping Wu, Liqun Huang, Julia Schnabel, Xiahai Zhuang
9 mins 9 secs
Abstract: Deep learning (DL)-based models have demonstrated good performance in medical image segmentation. However, the models trained on a known dataset often fail when performed on an unseen dataset collected from different centers, vendors and disease populations. In this work, we present a random style transfer network to tackle the domain generalization problem for multi-vendor and center cardiac image segmentation. Style transfer is used to generate training data with a wider distribution/ heterogeneity, namely domain augmentation. As the target domain could be unknown, we randomly generate a modality vector for the target modality in the style transfer stage, to simulate the domain shift for unknown domains. The model can be trained in a semi-supervised manner by simultaneously optimizing a supervised segmentation and a unsupervised style translation objective. Besides, the framework incorporates the spatial information and shape prior of the target by introducing two regularization terms. We evaluated the proposed framework on 40 subjects from the M&Ms challenge2020, and obtained promising performance in the segmentation for data from unknown vendors and centers.
Lei Li, Xin Weng, Julia Schnabel, Xiahai Zhuang
8 mins 41 secs
Abstract: We propose an end-to-end deep neural network (DNN) which can simultaneously segment the left atrial (LA) cavity and quantify LA scars. The framework incorporates the continuous spatial information of the target by introducing a spatially encoded (SE) loss based on the distance transform map. Compared to conventional binary label based loss, the proposed SE loss can reduce noisy patches in the resulting segmentation, which is commonly seen for deep learning-based methods. To fully utilize the inherent spatial relationship between LA and LA scars, we further propose a shape attention (SA) mechanism through an explicit surface projection to build an end-to-end-trainable model. Specifically, the SA scheme is embedded into a two-task network to perform the joint LA segmentation and scar quantification. Moreover, the proposed
method can alleviate the severe class-imbalance problem when detecting small and discrete targets like scars. We evaluated the proposed framework on 60 LGE MRI data from the MICCAI2018 LA challenge. For LA segmentation, the proposed method reduced the mean Hausdorff distance from 36.4mm to 20.0mm compared to the 3D basic U-Net using the binary cross-entropy loss. For scar quantification, the method was
compared with the results or algorithms reported in the literature and demonstrated better performance.
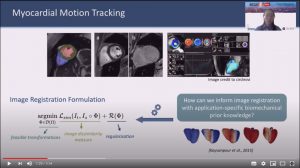
Biomechanics-informed Neural Network for Myocardial Motion Tracking in MRI
Chen Qin, Shuo Wang, Chen Chen, Huaqi Qiu, Wenjia Bai and Daniel Rueckert
9 mins 55 secs
Abstract: Image registration is an ill-posed inverse problem which often requires regularisation on the solution space. In contrast to most of the current approaches which impose explicit regularisation terms such as smoothness, in this paper we propose a novel method that can implicitly learn biomechanics-informed regularisation. Such an approach can incorporate application-specific prior knowledge into deep learning based registration. Particularly, the proposed biomechanics-informed regularisation leverages a variational autoencoder (VAE) to learn a manifold for biomechanically plausible deformations and to implicitly capture their underlying properties via reconstructing biomechanical simulations. The learnt VAE regulariser then can be coupled with any deep learning based
registration network to regularise the solution space to be biomechanically plausible. The proposed method is validated in the context of myocardial motion tracking on 2D stacks of cardiac MRI data from two different datasets. The results show that it can achieve better performance against other competing methods in terms of motion tracking accuracy and has the ability to learn biomechanical properties such as incompressibility and strains. The method has also been shown to have better generalisability to unseen domains compared with commonly used L2 regularisation schemes.
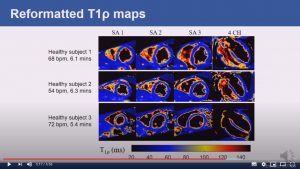
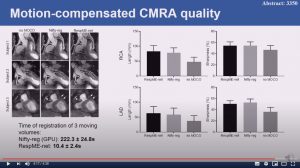
Respiratory motion-compensated high-resolution 3D whole-heart T1p mapping
Haikun Qi, Aurelien Bustin, Thomas Kuestner, Reza Hojhosseiny, Gastao Cruz, Karl Kunze, Radhouene Neji, Rene Botnar and Claudia Prieto
6 mins 59 secs
Abstract: Cardiovascular magnetic resonance (CMR) T1ρ mapping can be used to detect ischemic or non-ischemic cardiomyopathy without the need of exogenous contrast agents. Current 2D myocardial T1ρ mapping requires multiple breath-holds and provides limited coverage. Respiratory gating by diaphragmatic navigation has recently been exploited to enable free-breathing 3D T1ρ mapping, which, however, has low acquisition efficiency and may result in unpredictable and long scan times. This study aims to develop a fast respiratory motion-compensated 3D whole-heart myocardial T1ρ mapping technique with high spatial resolution and predictable scan time.
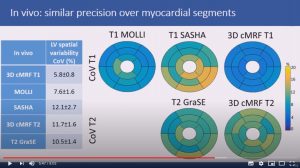
3D free-breathing cardiac magnetic resonance fingerprinting
Gastao Cruz, Olivier Jaubert, Haikun Qi, Aurelien Bustin, Giorgia Milotta, Torben Schneider, Peter Koken, Mariya Doneva, Rene Botnar and Claudia Prieto
8 mins 4 secs
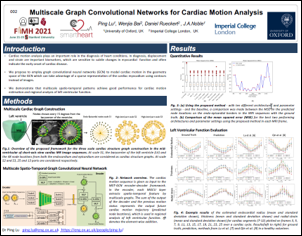
Ping Lu, Wenjia Bai, Daniel Rueckert and Alison Noble
9 mins 15 secs
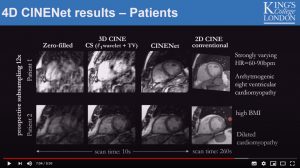
CINENet: Deep learning-based 3D Cardiac Cine Reconstruction with multi-coil complex 4D Spatio-Temporal Convolutions
Thomas Kuestner, Niccolo Fuin, Kerstin Hammernik, Radhouene Neji, Daniel Ruecker, Rene Botnar and Claudia Prieto
8 mins 59 secs
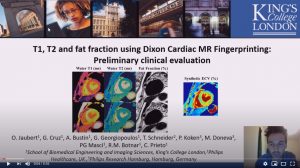
T1, T2 and fat fraction using Dixon Cardiac MR Fingerprinting: Preliminary clinical evaluation
Olivier Jaubert, G. Cruz, A. Bustin, G. Georgiopoulos, T. Schneider, P. Koken, M. Doneva, P.G. Masci, R.M. Botnar and C. Prieto
8 mins 37 secs
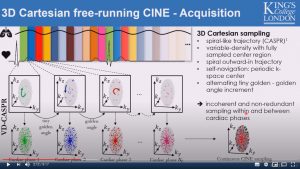
Fully self-gated free-breathing 3D Cartesian cardiac CINE with isotropic whole-heart coverage in less than 2 minutes
Thomas Kuestner, Aurelien Bustin, Olivier Jaubert, Rahouene Neji, Claudia Prieto and Rene Botnar
8 min 17 secs
Non-rigid bin-to-bin motion estimation of coronary MR images using unsupervised fully convolutional neural network
Haikun Qi, Gastao Cruz, Thomas Kuestner, Niccolo Fuin, Rene Botnar and Claudia Prieto
4 mins 36 secs