Tackling Big Data Challenges
My research center around four major directions:
Large-Scale Scientific Data/Spatial Data
My current research addresses developing novel data management algorithms for querying and analyzing big scientific data. The unprecedented size and growth of scientific datasets makes analyzing them a challenging data management problem. Current algorithms are not efficient for today’s data and will not scale to analyze the rapidly growing datasets of the future. I therefore want to develop next generation data management tools and techniques able to manage tomorrow’s scientific data, thereby putting efficient and scalable big data analytics at the fingertips of scientists so they can again focus on their science, unperturbed by the challenges of big data.
Key to my research is that the algorithms and methods developed are inspired by real use cases from other sciences and that they are also implemented and put to use for scientists. The algorithms developed so far are inspired by a collaboration with the neuroscientists from the Blue Brain project (BBP) who attempt to simulate the human brain on a supercomputer.
Large-Scale Data Analytics
Analysing massive amounts of data to extract value has become key across different disciplines. As the amounts of data grow rapidly, however, current approaches for data analysis struggle. I am consequently interested in developing new methods for the large-scale analysis of high-dimensional data. One direction is to do high-performance data analytics (HPDA) on supercomputing infrastructure. To do so existing algorithms need to be fundamentally redesigned and implemented to make use of the massive parallelism large-scale supercomputing infrastructure provide, i.e., analytics problems need to be formulated as embarrassingly parallel problems with little need for synchronization and communication. A second direction is to introduce approximation to enable analyses at scale. Introducing minimal approximation can accelerate analyses by several orders of magnitude and the goal of the research in this direction therefore is the development of new approximate analytics algorithms with tight error bounds.
Data Management on Novel Hardware
Hardware underlying data processing and analysis evolves at a rapid pace. New hardware offers interesting trade-offs which enables new data analysis techniques. Novel algorithms have to be developed for new storage technologies like Flash, PCM, 3D memory or even just abundantly available main memory. Similarly, new algorithms have to be developed for new processing technology like neuromorphic hardware.
Novel Data Storage Technology/DNA Storage
The demand for data-driven decision making coupled with need to retain data to meet regulatory compliance requirements has resulted in a rapid increase in the amount of archival data stored by enterprises. As data generation rate far outpaces the rate of improvement in storage density of media like HDD and tape, researchers have started investigating new architectures and media types that can store such “cold”, infrequently accessed data at very low cost.
Synthetic DNA is one such storage media that has received some attention recently due to its high density and durability. Open questions to be researched are how to integrate DNA in the database storage hierarchy. More specifically, the following two questions need to be addressed: (i) how can database knowledge help optimize DNA encoding and decoding? and (ii) how can biochemical mechanisms used for DNA manipulation be used to perform in-vitro, near-data SQL query processing?
Research Highlights
OligoArchive: Using DNA in the DBMS Storage Hierarchy
The demand for data-driven decision making coupled with need to retain data to meet regulatory compliance requirements has resulted in a rapid increase in the amount of archival data stored by enterprises. As data generation rate far outpaces the rate of improvement in storage density of media like HDD and tape, researchers have started investigating new architectures and media types that can store such “cold”, infrequently accessed data at very low cost.
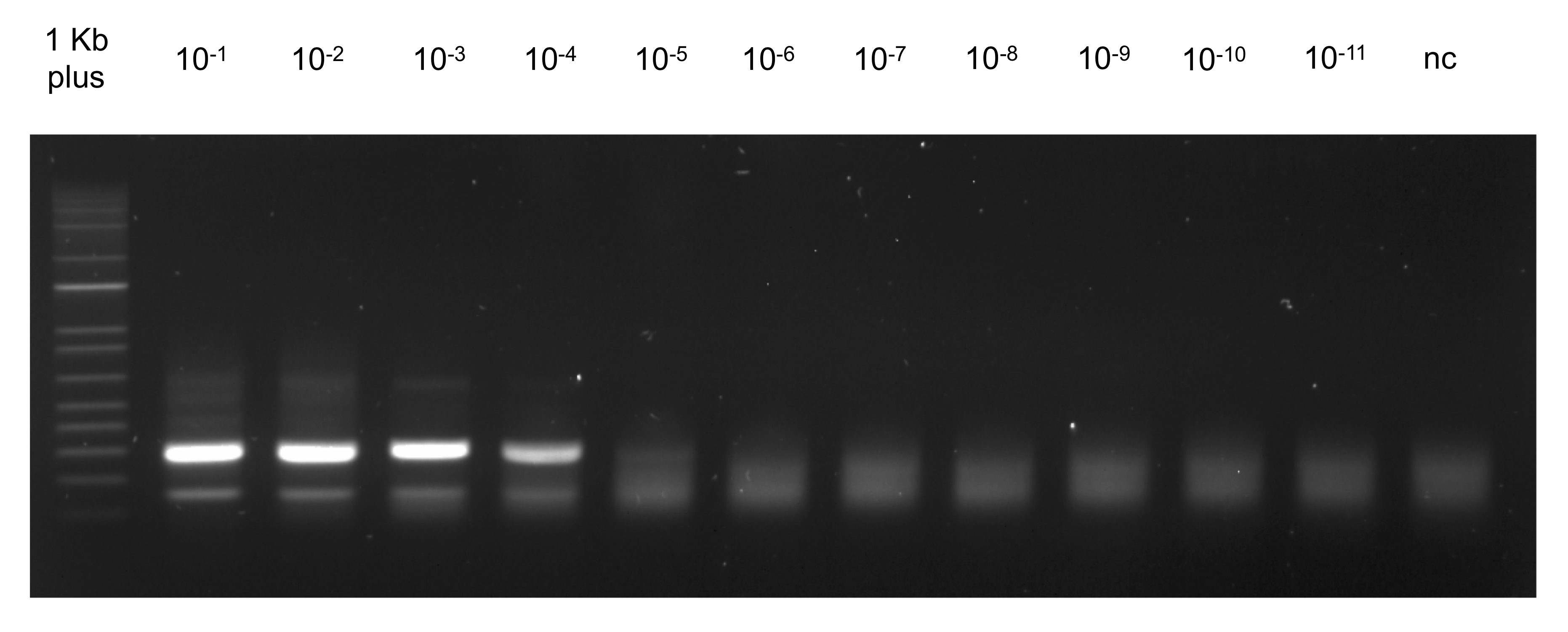
Synthetic DNA is one such storage media that has received some attention recently due to its high density and durability. In this paper, we investigate the problem of integrating DNA in the database storage hierarchy. More specifically, we ask the following two questions: (i) how can database knowledge help optimize DNA encoding and decoding? and (ii) how can biochemical mechanisms used for DNA manipulation be used to perform in-vitro, near-data SQL query processing?
In answering these questions, we present OligoArchive, an architecture for using DNA-based storage system as the archival tier of a relational database. We demonstrate that OligoArchive can be realized in practice by building archiving and recovery tools (pg_oligo_dump and pg_oligo_restore) for PostgreSQL that perform schema-aware encoding and decoding of relational data on DNA, and using these tools to archive a 12KB TPC-H database to DNA, perform in-vitro computation, and restore it back again. Find out more on the project website or in the paper
SCOUT: Prefetching for Latent Structure Following Queries
Today’s scientists are quickly moving from in vitro to in silico experimentation: they no longer analyze natural phenomena in a petri dish, but instead they build models and simulate them. Managing and analyzing the massive amounts of data involved in simulations is a major task. Yet, they lack the tools to efficiently work with data of this size.
One problem many scientists share is the analysis of the massive spatial models they build. For several types of analysis they need to interactively follow the structures in the spatial model, e.g., the arterial tree, neuron fibers, etc., and issue range queries along the way. Each query takes a long time to execute, and the total time for executing a sequence of queries significantly delays data analysis. Prefetching the spatial data reduces the response time considerably, but known approaches do not prefetch with high accuracy.
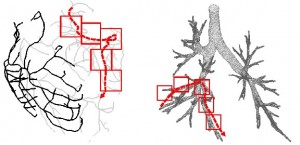
We have therefore developed SCOUT, a structure-aware method for prefetching data along interactive spatial query sequences. SCOUT uses an approximate graph model of the structures involved in past queries and attempts to identify what particular structure the user follows. Our experiments with neuroscience data show that SCOUT prefetches with an accuracy from 71% to 92%, which translates to a speedup of 4x-15x. SCOUT also improves the prefetching accuracy on datasets from other scientific domains, such as medicine and biology. Continue to read here…